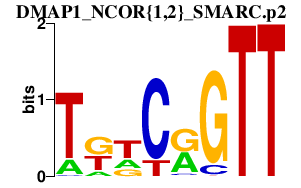
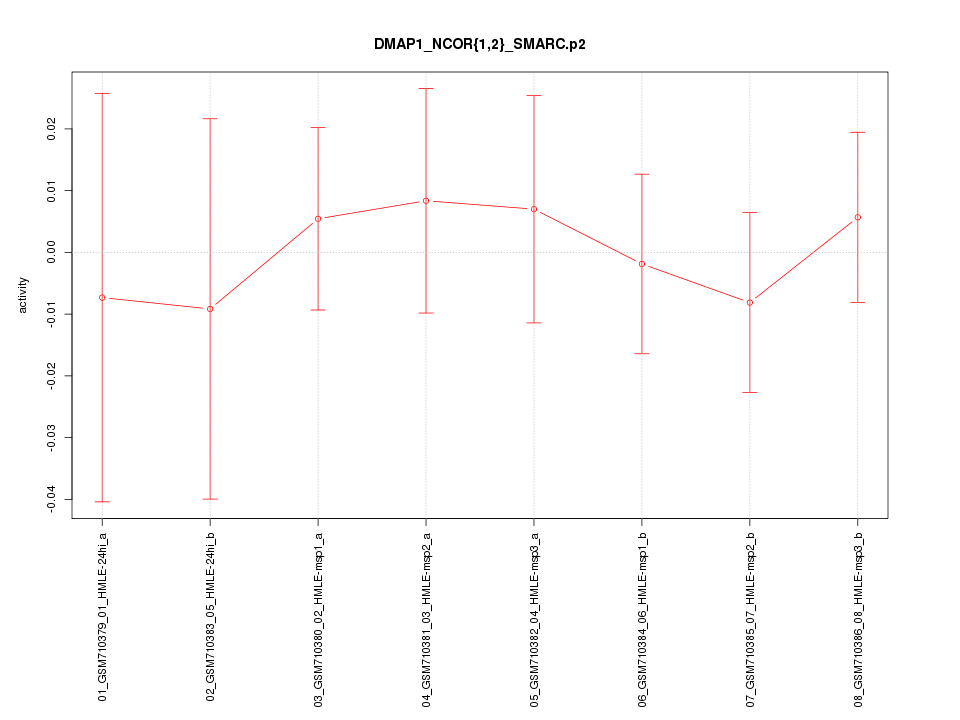
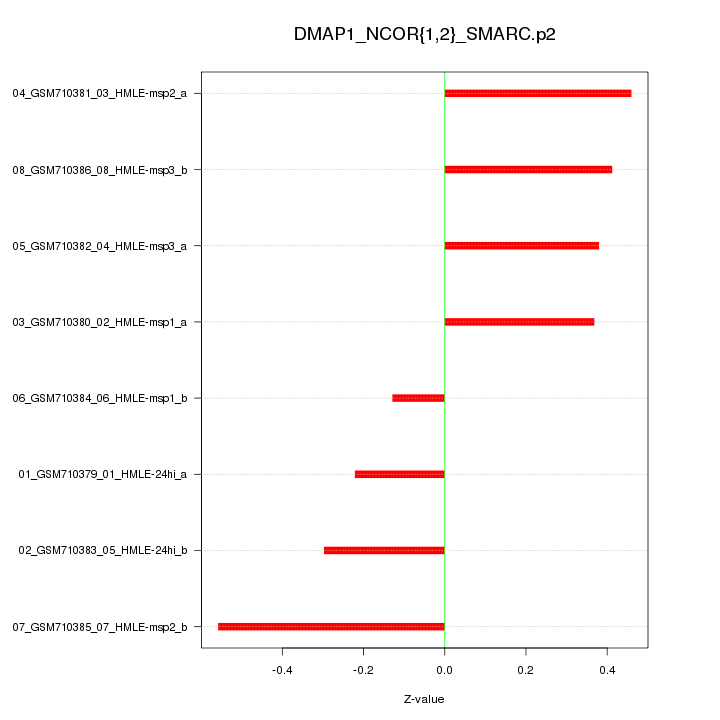
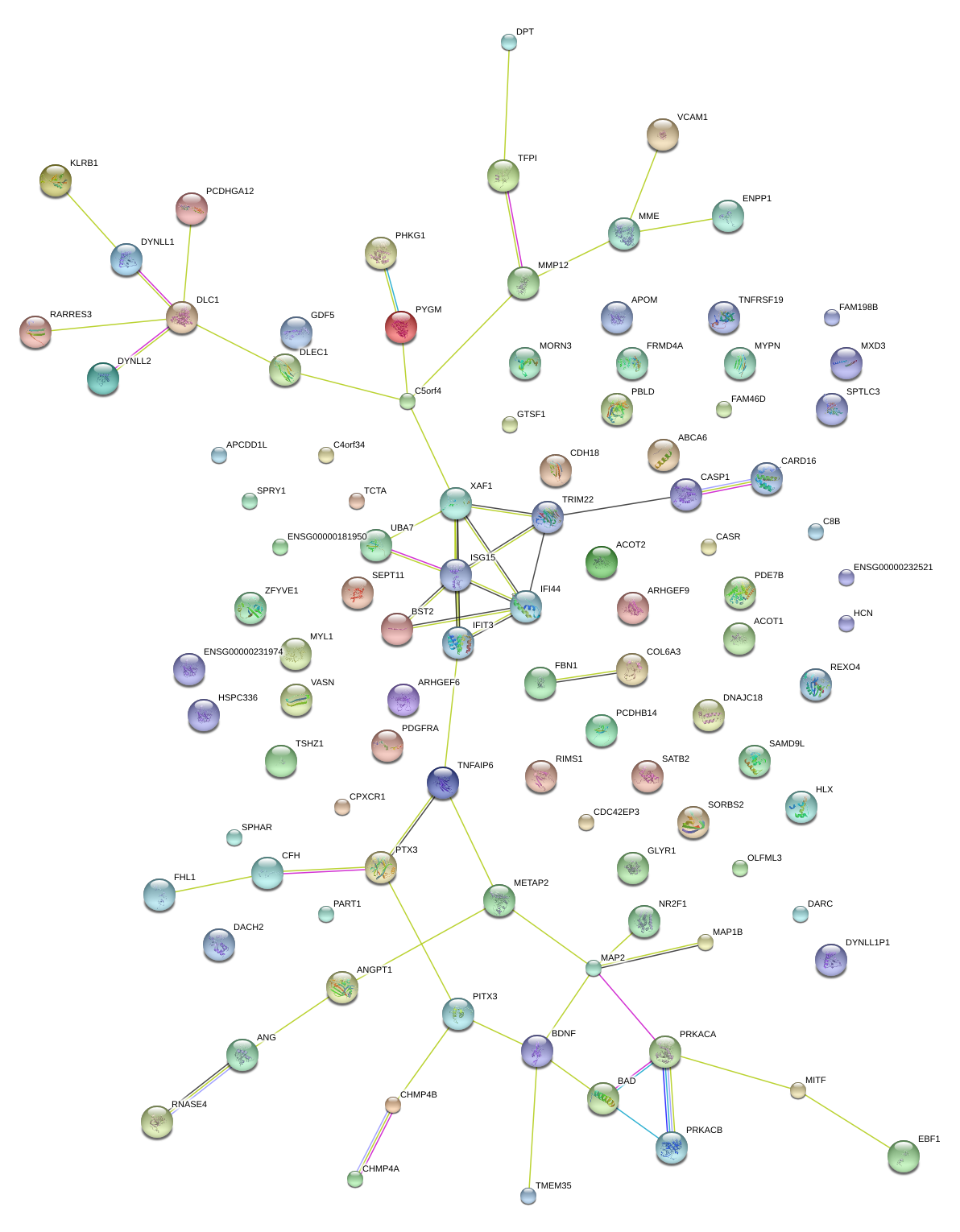
|
chr1_+_159175048
|
0.616
|
NM_001122951
|
DARC
|
Duffy blood group, chemokine receptor
|
|
chr5_+_92918924
|
0.424
|
NM_005654
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr8_-_12973752
|
0.318
|
NM_001164271
|
DLC1
|
deleted in liver cancer 1
|
|
chr1_-_168698412
|
0.293
|
NM_001937
|
DPT
|
dermatopontin
|
|
chr8_-_108510094
|
0.252
|
NM_001146
NM_001199859
|
ANGPT1
|
angiopoietin 1
|
|
chr5_-_19988293
|
0.237
|
NM_001167667
NM_004934
|
CDH18
|
cadherin 18, type 2
|
|
chr6_+_72596699
|
0.219
|
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr5_+_71502700
|
0.211
|
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr7_-_92777657
|
0.211
|
NM_152703
|
SAMD9L
|
sterile alpha motif domain containing 9-like
|
|
chr13_+_24144402
|
0.211
|
NM_001204459
NM_148957
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19
|
|
chr6_+_72596405
|
0.207
|
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr5_-_154230157
|
0.203
|
|
C5orf4
|
chromosome 5 open reading frame 4
|
|
chr5_-_154230212
|
0.194
|
NM_032385
|
C5orf4
|
chromosome 5 open reading frame 4
|
|
chr2_-_188419185
|
0.193
|
NM_001032281
NM_006287
|
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor)
|
|
chr1_+_221052735
|
0.188
|
NM_021958
|
HLX
|
H2.0-like homeobox
|
|
chr1_+_221052845
|
0.185
|
|
HLX
|
H2.0-like homeobox
|
|
chr16_-_4896197
|
0.182
|
|
GLYR1
|
glyoxylate reductase 1 homolog (Arabidopsis)
|
|
chr6_+_72922513
|
0.180
|
NM_001168407
NM_001168408
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr1_+_221053000
|
0.179
|
|
HLX
|
H2.0-like homeobox
|
|
chr6_+_136172802
|
0.166
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr20_-_34025955
|
0.161
|
NM_000557
|
GDF5
|
growth differentiation factor 5
|
|
chr1_+_114522029
|
0.161
|
NM_020190
|
OLFML3
|
olfactomedin-like 3
|
|
chr11_-_104916041
|
0.157
|
NM_001017534
NM_052889
|
CARD16
|
caspase recruitment domain family, member 16
|
|
chr17_+_6659356
|
0.157
|
|
XAF1
|
XIAP associated factor 1
|
|
chr2_-_188419049
|
0.155
|
|
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor)
|
|
chr6_+_31623668
|
0.146
|
NM_019101
|
APOM
|
apolipoprotein M
|
|
chr5_+_140602914
|
0.144
|
NM_018934
|
PCDHB14
|
protocadherin beta 14
|
|
chr12_-_54867372
|
0.143
|
NM_144594
|
GTSF1
|
gametocyte specific factor 1
|
|
chr11_+_63304272
|
0.141
|
NM_004585
|
RARRES3
|
retinoic acid receptor responder (tazarotene induced) 3
|
|
chr3_-_49851217
|
0.139
|
|
UBA7
|
ubiquitin-like modifier activating enzyme 7
|
|
chr4_-_39640461
|
0.134
|
NM_174921
|
C4orf34
|
chromosome 4 open reading frame 34
|
|
chr5_+_140810181
|
0.133
|
|
PCDHGA12
|
protocadherin gamma subfamily A, 12
|
|
chr2_+_152214105
|
0.133
|
NM_007115
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6
|
|
chr4_-_159094163
|
0.132
|
NM_001031700
NM_001128424
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr6_+_31623688
|
0.131
|
|
APOM
|
apolipoprotein M
|
|
chr19_-_17516383
|
0.129
|
NM_004335
|
BST2
|
bone marrow stromal cell antigen 2
|
|
chr6_-_138820580
|
0.129
|
|
|
|
|
chr6_+_72596648
|
0.124
|
NM_014989
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr2_+_210444154
|
0.124
|
|
MAP2
|
microtubule-associated protein 2
|
|
chr2_-_200325200
|
0.124
|
|
SATB2
|
SATB homeobox 2
|
|
chr11_+_5710926
|
0.123
|
|
TRIM22
|
tripartite motif containing 22
|
|
chr3_+_49449753
|
0.122
|
|
TCTA
|
T-cell leukemia translocation altered gene
|
|
chr14_+_21152735
|
0.121
|
|
ANG
RNASE4
|
angiogenin, ribonuclease, RNase A family, 5
ribonuclease, RNase A family, 4
|
|
chr3_+_69812961
|
0.118
|
NM_006722
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr5_+_59784004
|
0.117
|
|
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding)
|
|
chr6_+_132129195
|
0.116
|
|
ENPP1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1
|
|
chr1_+_79115476
|
0.116
|
NM_006417
|
IFI44
|
interferon-induced protein 44
|
|
chr11_-_64527369
|
0.115
|
|
PYGM
|
phosphorylase, glycogen, muscle
|
|
chr1_+_101185195
|
0.115
|
NM_001078
NM_001199834
NM_080682
|
VCAM1
|
vascular cell adhesion molecule 1
|
|
chr4_+_124317884
|
0.114
|
NM_199327
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila)
|
|
chr18_+_72922725
|
0.114
|
NM_005786
|
TSHZ1
|
teashirt zinc finger homeobox 1
|
|
chr10_+_69869249
|
0.112
|
NM_032578
|
MYPN
|
myopalladin
|
|
chr3_+_49449638
|
0.111
|
NM_022171
|
TCTA
|
T-cell leukemia translocation altered gene
|
|
chr5_-_138775138
|
0.111
|
|
DNAJC18
|
DnaJ (Hsp40) homolog, subfamily C, member 18
|
|
chr4_+_77953942
|
0.110
|
|
SEPT11
|
septin 11
|
|
chr2_-_37899227
|
0.110
|
NM_006449
|
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3
|
|
chr17_-_67138013
|
0.110
|
NM_080284
|
ABCA6
|
ATP-binding cassette, sub-family A (ABC1), member 6
|
|
chr20_+_12989616
|
0.105
|
NM_018327
|
SPTLC3
|
serine palmitoyltransferase, long chain base subunit 3
|
|
chr10_-_70066594
|
0.105
|
|
PBLD
|
phenazine biosynthesis-like protein domain containing
|
|
chr1_+_948846
|
0.104
|
NM_005101
|
ISG15
|
ISG15 ubiquitin-like modifier
|
|
chr10_-_14372807
|
0.104
|
NM_018027
|
FRMD4A
|
FERM domain containing 4A
|
|
chr11_+_65192284
|
0.102
|
|
NEAT1
|
nuclear paraspeckle assembly transcript 1 (non-protein coding)
|
|
chr1_-_57431620
|
0.102
|
NM_000066
|
C8B
|
complement component 8, beta polypeptide
|
|
chr3_+_157154579
|
0.100
|
NM_002852
|
PTX3
|
pentraxin 3, long
|
|
chr11_+_5710816
|
0.100
|
NM_001199573
NM_006074
|
TRIM22
|
tripartite motif containing 22
|
|
chr2_-_238322770
|
0.099
|
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr17_+_6659155
|
0.098
|
NM_017523
NM_199139
|
XAF1
|
XIAP associated factor 1
|
|
chr5_+_140810126
|
0.097
|
NM_003735
NM_032094
|
PCDHGA12
|
protocadherin gamma subfamily A, 12
|
|
chr4_+_55095263
|
0.096
|
NM_006206
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr11_+_65268313
|
0.096
|
|
MALAT1
|
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding)
|
|
chr2_-_211179765
|
0.095
|
NM_079420
|
MYL1
|
myosin, light chain 1, alkali; skeletal, fast
|
|
chr14_-_73493744
|
0.094
|
NM_021260
|
ZFYVE1
|
zinc finger, FYVE domain containing 1
|
|
chr14_+_21156922
|
0.094
|
NM_001097577
NM_194431
|
ANG
RNASE4
|
angiogenin, ribonuclease, RNase A family, 5
ribonuclease, RNase A family, 4
|
|
chr3_+_69812620
|
0.093
|
NM_001184967
|
MITF
|
microphthalmia-associated transcription factor
|
|
chrX_+_88002225
|
0.092
|
NM_001184771
NM_033048
|
CPXCR1
|
CPX chromosome region, candidate 1
|
|
chr14_-_24683035
|
0.091
|
NM_014169
|
CHMP4A
|
charged multivesicular body protein 4A
|
|
chr5_+_140753480
|
0.091
|
NM_018919
NM_032086
|
PCDHGA6
|
protocadherin gamma subfamily A, 6
|
|
chr1_+_229440128
|
0.091
|
NM_006542
|
SPHAR
|
S-phase response (cyclin related)
|
|
chrX_+_100333835
|
0.090
|
NM_021637
|
TMEM35
|
transmembrane protein 35
|
|
chrX_-_63005212
|
0.090
|
NM_001173479
|
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9
|
|
chr3_-_151987332
|
0.089
|
|
LOC401093
|
uncharacterized LOC401093
|
|
chr5_-_176738831
|
0.089
|
|
MXD3
|
MAX dimerization protein 3
|
|
chr6_+_73076431
|
0.087
|
NM_001168411
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr5_-_158526698
|
0.084
|
NM_024007
|
EBF1
|
early B-cell factor 1
|
|
chr1_+_84609941
|
0.083
|
NM_182948
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr11_-_64052164
|
0.083
|
NM_004322
NM_032989
|
BAD
|
BCL2-associated agonist of cell death
|
|
chrX_+_79675945
|
0.081
|
NM_152630
|
FAM46D
|
family with sequence similarity 46, member D
|
|
chrX_-_135863033
|
0.081
|
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr11_-_2018568
|
0.080
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr4_-_186733372
|
0.080
|
NM_003603
NM_001145670
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr3_+_154798078
|
0.080
|
NM_007289
|
MME
|
membrane metallo-endopeptidase
|
|
chr20_-_57089845
|
0.080
|
NM_153360
|
APCDD1L
|
adenomatosis polyposis coli down-regulated 1-like
|
|
chrX_+_135251795
|
0.080
|
NM_001167819
|
FHL1
|
four and a half LIM domains 1
|
|
chr14_+_74003817
|
0.080
|
NM_001037161
|
ACOT1
|
acyl-CoA thioesterase 1
|
|
chr10_+_91092238
|
0.079
|
NM_001031683
|
IFIT3
|
interferon-induced protein with tetratricopeptide repeats 3
|
|
chr15_-_48937795
|
0.079
|
NM_000138
|
FBN1
|
fibrillin 1
|
|
chr5_-_138775213
|
0.079
|
NM_152686
|
DNAJC18
|
DnaJ (Hsp40) homolog, subfamily C, member 18
|
|
chrX_+_85517970
|
0.079
|
NM_001139515
|
DACH2
|
dachshund homolog 2 (Drosophila)
|
|
chr16_+_4430878
|
0.078
|
|
VASN
|
vasorin
|
|
chr11_-_27681195
|
0.077
|
NM_001143816
NM_170735
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr3_-_100712248
|
0.077
|
NM_015429
|
ABI3BP
|
ABI family, member 3 (NESH) binding protein
|
|
chr3_+_119316694
|
0.076
|
NM_001206960
NM_001206961
NM_015900
|
PLA1A
|
phospholipase A1 member A
|
|
chr3_+_45927995
|
0.076
|
NM_006641
NM_031200
|
CCR9
|
chemokine (C-C motif) receptor 9
|
|
chrX_+_80457559
|
0.076
|
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like
|
|
chr5_+_133842276
|
0.076
|
|
|
|
|
chrX_-_18372777
|
0.075
|
NM_006089
|
SCML2
|
sex comb on midleg-like 2 (Drosophila)
|
|
chr1_-_163172624
|
0.074
|
NM_001195303
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr12_-_10324675
|
0.074
|
NM_001172632
NM_001172633
NM_002543
|
OLR1
|
oxidized low density lipoprotein (lectin-like) receptor 1
|
|
chr1_-_151319717
|
0.073
|
NM_000449
NM_001025603
|
RFX5
|
regulatory factor X, 5 (influences HLA class II expression)
|
|
chr2_-_214015053
|
0.072
|
NM_016260
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr19_+_17516502
|
0.072
|
|
LOC100507535
|
uncharacterized LOC100507535
|
|
chr4_-_159092509
|
0.071
|
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr13_+_49550949
|
0.071
|
|
FNDC3A
|
fibronectin type III domain containing 3A
|
|
chr2_-_68547018
|
0.071
|
NM_001111101
NM_015463
|
CNRIP1
|
cannabinoid receptor interacting protein 1
|
|
chr3_-_45267068
|
0.070
|
|
TMEM158
|
transmembrane protein 158 (gene/pseudogene)
|
|
chr4_-_57547845
|
0.070
|
NM_001145460
NM_032495
NM_139212
|
HOPX
|
HOP homeobox
|
|
chr18_-_3011944
|
0.070
|
NM_014646
|
LPIN2
|
lipin 2
|
|
chr8_-_93029864
|
0.070
|
NM_175636
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr2_-_167232464
|
0.069
|
NM_002977
|
SCN9A
|
sodium channel, voltage-gated, type IX, alpha subunit
|
|
chr10_+_91152321
|
0.069
|
NM_001548
|
IFIT1
|
interferon-induced protein with tetratricopeptide repeats 1
|
|
chr1_-_151319674
|
0.068
|
|
RFX5
|
regulatory factor X, 5 (influences HLA class II expression)
|
|
chr16_+_15528324
|
0.068
|
NM_033201
|
C16orf45
|
chromosome 16 open reading frame 45
|
|
chr8_-_60031545
|
0.068
|
NM_014729
|
TOX
|
thymocyte selection-associated high mobility group box
|
|
chr17_-_8702438
|
0.068
|
NM_152599
|
MFSD6L
|
major facilitator superfamily domain containing 6-like
|
|
chr19_+_10197023
|
0.067
|
|
C19orf66
|
chromosome 19 open reading frame 66
|
|
chr3_+_159557745
|
0.067
|
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr2_+_210444364
|
0.067
|
NM_002374
NM_031845
NM_031847
|
MAP2
|
microtubule-associated protein 2
|
|
chr19_+_10196972
|
0.066
|
|
C19orf66
|
chromosome 19 open reading frame 66
|
|
chr17_-_29641068
|
0.066
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr15_+_97326629
|
0.065
|
NM_173499
|
SPATA8
|
spermatogenesis associated 8
|
|
chr16_+_8814567
|
0.064
|
NM_001127448
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr11_-_64052123
|
0.064
|
|
BAD
|
BCL2-associated agonist of cell death
|
|
chr3_+_187086167
|
0.063
|
NM_022147
|
RTP4
|
receptor (chemosensory) transporter protein 4
|
|
chr6_+_25962978
|
0.063
|
NM_006355
|
TRIM38
|
tripartite motif containing 38
|
|
chr6_-_167571315
|
0.062
|
NM_005299
|
GPR31
|
G protein-coupled receptor 31
|
|
chr3_-_49851356
|
0.062
|
NM_003335
|
UBA7
|
ubiquitin-like modifier activating enzyme 7
|
|
chr19_-_10679328
|
0.062
|
|
CDKN2D
|
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4)
|
|
chr1_+_162602308
|
0.061
|
|
DDR2
|
discoidin domain receptor tyrosine kinase 2
|
|
chr4_-_186732208
|
0.061
|
NM_001145671
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr4_+_74347461
|
0.061
|
NM_001133
|
AFM
|
afamin
|
|
chr4_-_159093717
|
0.060
|
NM_016613
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr9_-_73029539
|
0.060
|
NM_001206
|
KLF9
|
Kruppel-like factor 9
|
|
chr14_+_76618272
|
0.060
|
|
C14orf118
|
chromosome 14 open reading frame 118
|
|
chr5_-_169724614
|
0.059
|
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr5_-_138775189
|
0.059
|
|
DNAJC18
|
DnaJ (Hsp40) homolog, subfamily C, member 18
|
|
chr6_+_25963398
|
0.059
|
|
TRIM38
|
tripartite motif containing 38
|
|
chr1_-_113161724
|
0.059
|
NM_017744
NM_138727
NM_138728
NM_138729
|
ST7L
|
suppression of tumorigenicity 7 like
|
|
chr11_-_2019007
|
0.059
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chrX_+_9431314
|
0.058
|
NM_001139466
NM_001139467
NM_001139468
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr3_+_29322802
|
0.058
|
NM_001003792
NM_001003793
NM_001177711
NM_001177712
NM_014483
|
RBMS3
|
RNA binding motif, single stranded interacting protein 3
|
|
chr1_+_202431870
|
0.058
|
NM_032103
NM_032104
|
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B
|
|
chr18_-_25756946
|
0.058
|
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal)
|
|
chr1_+_162602198
|
0.057
|
NM_001014796
NM_006182
|
DDR2
|
discoidin domain receptor tyrosine kinase 2
|
|
chr10_-_74283693
|
0.057
|
NM_001195519
|
MICU1
|
mitochondrial calcium uptake 1
|
|
chr16_+_55600583
|
0.057
|
NM_032330
|
CAPNS2
|
calpain, small subunit 2
|
|
chr19_-_44384240
|
0.056
|
NM_001033719
|
ZNF404
|
zinc finger protein 404
|
|
chr3_-_50374875
|
0.056
|
NM_170713
|
RASSF1
|
Ras association (RalGDS/AF-6) domain family member 1
|
|
chr8_+_17396285
|
0.056
|
NM_001164771
NM_003046
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr3_-_122283423
|
0.056
|
NM_001146103
NM_031458
|
PARP9
|
poly (ADP-ribose) polymerase family, member 9
|
|
chr6_+_44126547
|
0.056
|
NM_007058
|
CAPN11
|
calpain 11
|
|
chr20_-_39946119
|
0.056
|
|
ZHX3
|
zinc fingers and homeoboxes 3
|
|
chr3_+_49059046
|
0.056
|
NM_199069
NM_199070
|
NDUFAF3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 3
|
|
chrX_-_106960268
|
0.055
|
NM_004089
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr3_-_169587598
|
0.054
|
NM_024727
|
LRRC31
|
leucine rich repeat containing 31
|
|
chr12_-_7245006
|
0.054
|
NM_001733
|
C1R
|
complement component 1, r subcomponent
|
|
chr1_-_46598250
|
0.054
|
NM_003629
NM_001114172
|
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma)
|
|
chr3_+_159557649
|
0.054
|
NM_001197109
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chrX_+_110366304
|
0.054
|
NM_001128168
NM_001128172
NM_001128173
|
PAK3
|
p21 protein (Cdc42/Rac)-activated kinase 3
|
|
chr1_+_244998592
|
0.054
|
NM_198076
|
FAM36A
|
family with sequence similarity 36, member A
|
|
chr13_+_49550911
|
0.053
|
|
FNDC3A
|
fibronectin type III domain containing 3A
|
|
chr2_-_230579198
|
0.053
|
NM_139072
|
DNER
|
delta/notch-like EGF repeat containing
|
|
chr19_-_18508407
|
0.053
|
NM_145256
|
LRRC25
|
leucine rich repeat containing 25
|
|
chr1_-_113162215
|
0.052
|
|
ST7L
|
suppression of tumorigenicity 7 like
|
|
chr11_-_102595623
|
0.052
|
NM_002424
|
MMP8
|
matrix metallopeptidase 8 (neutrophil collagenase)
|
|
chr1_-_173886401
|
0.052
|
|
SERPINC1
|
serpin peptidase inhibitor, clade C (antithrombin), member 1
|
|
chr9_-_21202201
|
0.051
|
NM_021057
|
IFNA7
|
interferon, alpha 7
|
|
chr6_+_31913708
|
0.051
|
NM_001710
|
CFB
|
complement factor B
|
|
chr13_+_49822046
|
0.051
|
NM_001193478
NM_030911
|
CDADC1
|
cytidine and dCMP deaminase domain containing 1
|
|
chr10_-_44063906
|
0.051
|
NM_005674
|
ZNF239
|
zinc finger protein 239
|
|
chr6_+_132129154
|
0.051
|
NM_006208
|
ENPP1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1
|
|
chr7_+_100728719
|
0.051
|
NM_030961
|
TRIM56
|
tripartite motif containing 56
|
|
chr17_-_67311773
|
0.051
|
NM_018672
|
ABCA5
|
ATP-binding cassette, sub-family A (ABC1), member 5
|
|
chr20_-_35488275
|
0.050
|
NM_199181
|
KIAA0889
|
KIAA0889
|
|
chr4_+_41614725
|
0.050
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chrX_-_135863104
|
0.050
|
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr12_-_46357949
|
0.050
|
|
|
|
|
chr6_+_108881010
|
0.050
|
NM_201559
|
FOXO3
|
forkhead box O3
|
|
chr6_+_31913763
|
0.050
|
|
CFB
|
complement factor B
|
|
chr17_-_39183453
|
0.049
|
NM_031957
|
KRTAP1-5
|
keratin associated protein 1-5
|
|
chr1_+_145096391
|
0.049
|
NM_004892
|
SEC22B
|
SEC22 vesicle trafficking protein homolog B (S. cerevisiae) (gene/pseudogene)
|
|
chr6_-_48078909
|
0.049
|
|
C6orf138
|
chromosome 6 open reading frame 138
|
|
chr8_-_87520849
|
0.049
|
|
FAM82B
|
family with sequence similarity 82, member B
|
|
chr1_+_51434375
|
0.049
|
|
CDKN2C
|
cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4)
|
|
chr20_-_47894591
|
0.049
|
NM_021035
|
ZNFX1
|
zinc finger, NFX1-type containing 1
|
|
chr3_+_154797910
|
0.049
|
NM_007288
|
MME
|
membrane metallo-endopeptidase
|
|
chr11_-_2170792
|
0.048
|
NM_001007139
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr3_+_138066489
|
0.048
|
NM_001252091
NM_012219
|
MRAS
|
muscle RAS oncogene homolog
|
|
chr17_-_46688279
|
0.048
|
|
HOXB7
|
homeobox B7
|
|
chr20_+_34995443
|
0.047
|
NM_014902
|
DLGAP4
|
discs, large (Drosophila) homolog-associated protein 4
|
|
chr3_-_15839674
|
0.047
|
NM_001195098
|
ANKRD28
|
ankyrin repeat domain 28
|